Research
Our genomes are organized in a polymeric complex called chromatin with a repeating unit called the nucleosome. Long thought of as a repressive complex, it is now clear that the nucleosome serves as a vibrant signaling hub for genome-templated processes. We use structural biology, protein chemistry, and proteomics to better understand chromatin signaling in health and disease.
Determining the structural basis of histone modification cross-talk
A large part of our research program centers on using cryo-electron microscopy (cryo-EM) and X-ray crystallography to understand nucleosome recognition by chromatin enzymes at near atomic resolution. Our primary goal is to define mechanisms governing specificity, multivalency, and crosstalk in chromatin signaling. Through understanding how these enzymes function, we gain insight into their roles in genome-templated processes and their misregulation in disease, especially cancer. We are uniquely poised to study chromatin signaling due to our ability to build site-specifically modified nucleosomes using protein chemistry tools like expressed protein ligation. For example, we recently solved the cryo-EM structure of the histone H3 lysine methyltransferase Dot1L bound to a site-specifically, chemically ubiquitylated nucleosome. Dot1L requires prior ubiquitylation of histone H2B for optimal activity. Misregulation of this histone crosstalk pathway is pathogenic in a subset of leukemias and Dot1L inhibitors are now in clinical trials. Our structure shows how Dot1L binds its nucleosome substrate in a poised state and is activated by ubiquitin. This structure exemplifies the power of combining protein chemistry, biochemistry, and structural biology to understand mechanisms of chromatin signaling with atomic precision at near-atomic resolution. Building on our successes with Dot1L, our research program aims to solve many structures of chromatin enzymes bound to modified nucleosomes.
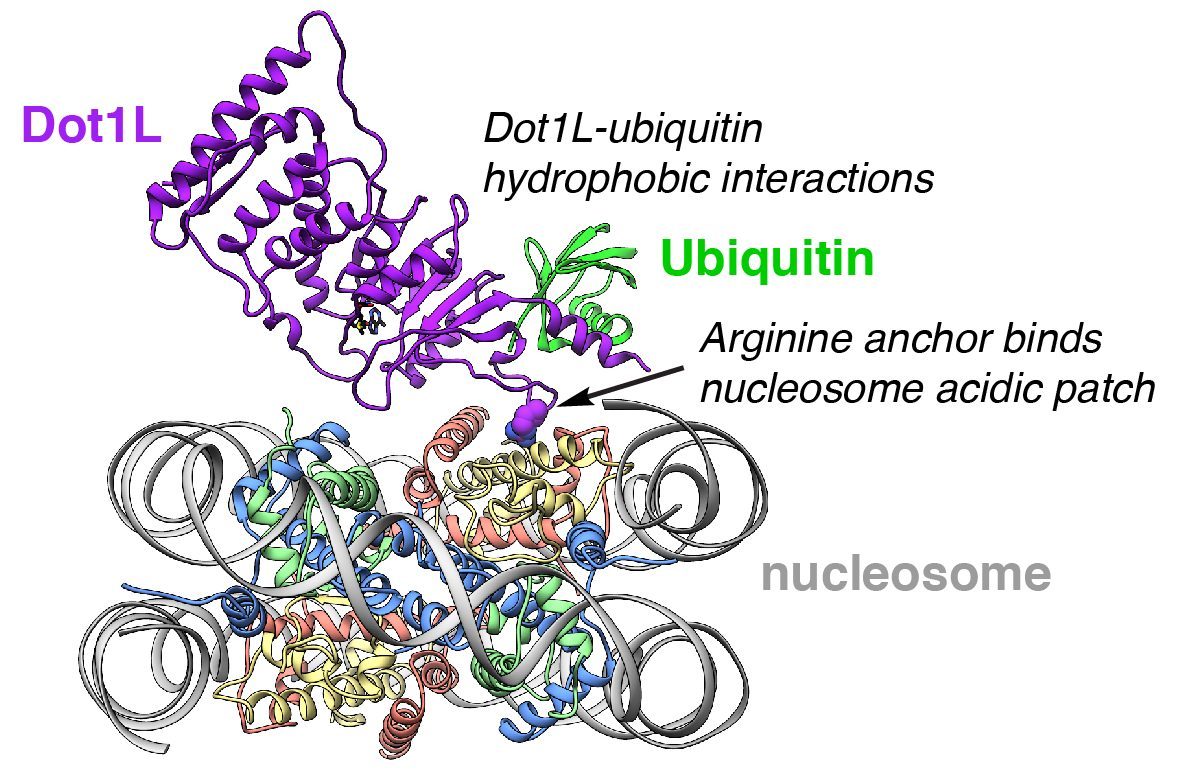
Cryo-EM structure of leukemogenic histone methyltransferase Dot1L bound to a ubiquitylated nucleosome substrate.
Establishing paradigms of nucleosome recognition
Current understanding of the patterns for nucleosome recognition by chromatin enzymes has been slowly and anecdotally built through structural characterization of individual chromatin enzyme-nucleosome complexes. Using these structures, we have proposed an emerging paradigm of nucleosome recognition in which arginines from the chromatin enzymes (that we have named arginine anchors) bind to an acidic patch on the nucleosome surface. However, the pervasiveness of the arginine anchor-nucleosome acidic patch interaction and existence of other paradigms for nucleosome recognition are unclear. We are using proteomics to define patterns of nucleosome recognition with the goal of establishing a universal framework for nucleosome binding.
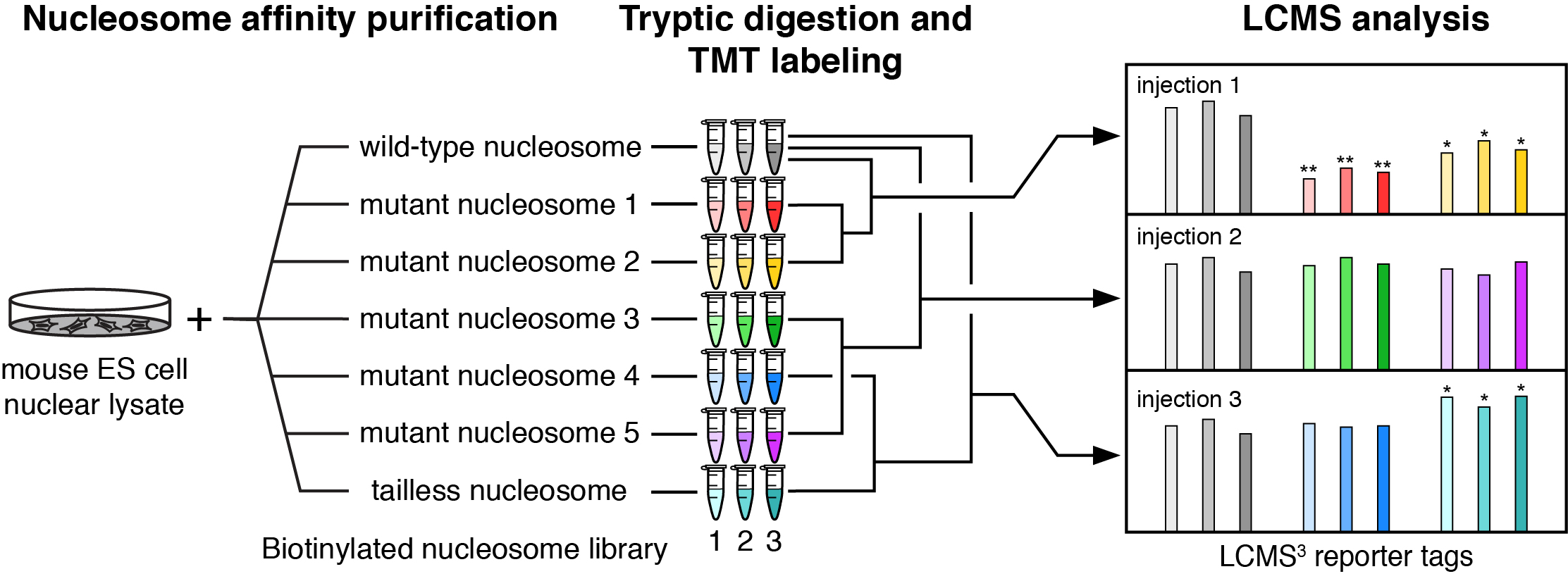
Workflow for nucleosome interactome proteomics.
Creating tools to enable epigenetics research
We also are developing new tools to probe epigenetic signaling both in vitro and in model systems. Such tools will allow precise hypothesis-driven interrogation of epigenetic signaling networks.
Our funding sources
![]()




![]()
